Matthew K Martz
PhD
Machine Learning · AI · Biochemistry · Molecular Biology · Cell Biology
- AI/Machine Learning and Algorithm Development - Penrose Hill, NYC
- Postdoctoral Fellow - Biochemistry and Biophysics, University of North Carolina
- PhD - Biochemistry and Molecular Biology, Thomas Jefferson University
- BS - Biochemistry and Molecular Biology, Pennsylvania State University
AI and Machine Learning Research
Updating this content to discuss current research direction. Please reach out
with to discuss in the meantime.
Biomedical Sciences
Cell biology, a centuries old field dating back to the early work of Robert
Hooke in his observations of plant cell walls[1] and Antonie van Leeuwenhoek with his analysis of
'living cells'[2], is the study of how
and why cells function as they do. My work and interests lie in uncovering the
connection between low level chemical processes and the high level cellular
events that epitomize these fundamental units of life. Elucidating how this
information is initiated and compiled reveals an understanding of normal
cellular function while introducing questions as to how perturbations to these
intrinsic processes are handled.
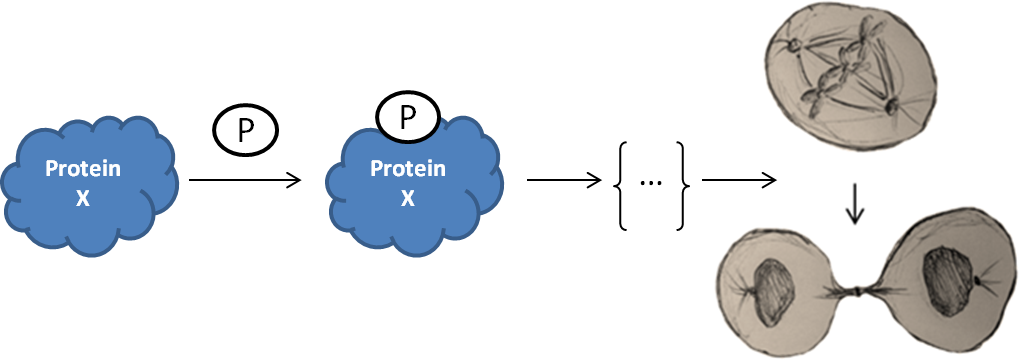 In addressing questions of this sort, research evolves as a sort of biphasic
process. The first is the establishment of signaling pathways in the cell in
both the normal state and in response to environmental stimuli, and how these
lead to physiological changes. The second is how the cell detects and handles
aberrations to these processes to ensure fidelity. However, both phases require
the pursuit of a similar root understanding. Whether interrogating a typical
housekeeping pathway or a regulatory network attenuating an aberrant signal, the
cell must translate low level chemical changes to high level outcomes. It is not
surprising that cell biology is growing to encompass numerous fields of study
and exemplifies the emergence of truly multidisciplinary research.
In addressing questions of this sort, research evolves as a sort of biphasic
process. The first is the establishment of signaling pathways in the cell in
both the normal state and in response to environmental stimuli, and how these
lead to physiological changes. The second is how the cell detects and handles
aberrations to these processes to ensure fidelity. However, both phases require
the pursuit of a similar root understanding. Whether interrogating a typical
housekeeping pathway or a regulatory network attenuating an aberrant signal, the
cell must translate low level chemical changes to high level outcomes. It is not
surprising that cell biology is growing to encompass numerous fields of study
and exemplifies the emergence of truly multidisciplinary research.
Research Interests
- fundamental questions of cellular systems with a particular interest in cell signaling
- understanding fate-based decision processes during complex cellular dynamics and how regulatory networks handle aberrations
- how low-level chemical processes translate into high-level decision making in cells
- integrative approaches to asking scientific questions combining traditional biochemical and molecular biological techniques with novel microscopy methods
- translating cellular systems to computational machines to ask questions from an algorithmic standpoint
- adaption of machine learning techniques in the analysis of experimental data and in model development
Systems biology approaches these complex interactions that manifest in
high level outcomes from a holistic perspective and in doing so, I aim
to adopt an effective integrative approach in answering cell
biological questions. Utilization of both traditional biochemical and
molecular biological techniques with cross-disciplinary computational
methodologies not only allows for ways to manage and interpret data on
previously unachievable scales, it provides platforms for asking
question and addressing questions in novel ways. This is exemplified
in the ability to analyze derivations in, or interactions between,
vast regulatory networks. However, the adoption of computer and data
sciences is not limited simply to the large scale gathering, storing,
and analysis of experimental data, these fields can allow for exciting
new theoretical ds escriptions of cells.
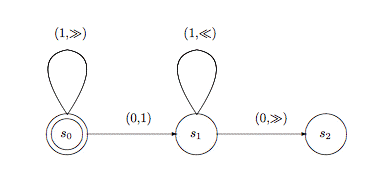 Addressing problems from a systems level requires an effective integrative
research approach, combining traditional biochemical and molecular biological
techniques with novel computational and microscopy methodologies. While adapting
new technologies for basic science research I will be further working towards
developing models of cellular processes and their regulation as computational
machines. The goal is to be able to establish the basic algorithms of these
networks and develop sets of cellular heuristics that comprise the decisions
that are undertaken. This algorithmic description of cells will further enhance
our understanding of cell biology while allowing for novel quantitative
approaches from a systems level.
Addressing problems from a systems level requires an effective integrative
research approach, combining traditional biochemical and molecular biological
techniques with novel computational and microscopy methodologies. While adapting
new technologies for basic science research I will be further working towards
developing models of cellular processes and their regulation as computational
machines. The goal is to be able to establish the basic algorithms of these
networks and develop sets of cellular heuristics that comprise the decisions
that are undertaken. This algorithmic description of cells will further enhance
our understanding of cell biology while allowing for novel quantitative
approaches from a systems level.
I may be reached most readily via email
Selected recent publications:
-
Ramona Schrage, ..., Matthew Martz, ..., Evi Kostenis. The experimental power of FR900359 to study Gq-regulated biological processes. Nature Communications 6, Article number: 10156. 14 December 2015.
-
Michelle C Helms, Elda Grabocka, Matthew K Martz, Christopher C Fischer, Nobuchika Suzuki, Philip B Wedegaertner. Mitotic-dependent phosphorylation of leukemia-associated RhoGEF (LARG) by Cdk1. Cellular Signalling, Volume 28, Issue 1, January 2016, Pages 43-52.
-
Martz MK, Grabocka E, Beeharry N, Yen TJ, Wedegaertner PW. Leukemia-Associated RhoGEF (LARG) is a Novel RhoGEF in Cytokinesis and Required for the Proper Completion of Abscission. Mol. Biol. Cell September 15, 2013 vol. 24 no. 18 2785-2794.
This paper describes a novel RhoGEF in mitosis acting at a
temporally distinct point in the cytokinetic process. We
demonstrate that LARG is required for proper abscission kinetics
in membrane scission between nascent forming daughter cells. The
depletion of LARG from cells further appears to result in
cytokinetic apoptosis. Moreover, we establish the mitotic kinase
Aurora B as mediator of perturbed abscission kinetics in the
abscence of LARG and highlights a more omnipotent abscission
checkpoint. Together with recent work in late cytokinesis, these
studies reveal a more complex role for RhoA signaling
post-contractile ring ingrestion.
-
Matthew Martz and Philip Wedegaertner: Faculty of 1000 Biology, 23 Jul 2010 F1000Prime.com/4242964#eval4039063
A commentary on a wonderful paper from the Siderovski lab
demonstrating support for a kinetic scaffold model wherein the GTP
hydrolysis activity of RGS proteins alone is sufficient to confer
both signal onset and decay.
-
Carkaci-Salli N, Flanagan JM, Martz MK, Salli U, Walther DJ, Bader M, Vrana KE. Functional domains of human tryptophan hydroxylase 2 (hTPH2). J Biol Chem. 2006 Sep 22;281(38):28105-12. Epub 2006 Jul 24.
This work begins to indentify the structure-function
relationship of hTPH2, the central nervous system isoform of the
enzyme responsible for the rate limiting, initial step in
serotonin production. We describe the purification and
characterization of both the wildtype enzyme as and several
important truncation mutants in terms of protein stability and
enzyme function. These procedural studies should prove invaluable
in future kinetic studies of hTPH2 as well as highlight key
mutants in obtaining stable protein for structural
analysis.
Relevant publication information can be found at Google Scholar
Selected recent talks and presentations:
- 2013 Invited Participant American Chemical Society Summer Institute
- Washington, DC
- 2012 Poster American Society for Cell Biology Meeting
- San Francisco, CA
- 2012 Invited Poster Lennox K. Black International Prize for Excellence in Biomedical Research 7th Symposium
- Thomas Jefferson University
- 2012 Talk Biochemistry Department Retreat
- Thomas Jefferson University
- 2012 Poster Experimental Biology
- San Diego, CA
- 2011 Judge Sigma Xi Research Day
- Thomas Jefferson University
- 2011 Poster Mid Atlantic Pharmacology Society Meeting
- Thomas Jefferson University
- 2011 Talk Research in Progress
- Thomas Jefferson University
- 2011 Gordon Research Conference on Molecular Pharmacology
- Ventura, CA
- 2010 Poster Sigma Xi Research Day
- Thomas Jefferson University
- 2010 Poster G Protein Signaling Workshop
- Philadelphia, PA
- 2010 Poster Biochemistry Department Retreat
- Thomas Jefferson University
- 2010 American Society for Cell Biology Meeting
- Philadelphia, PA
- 2010 Talk Research in Progress
- Thomas Jefferson University
- 2008 Poster Sigma Xi Research Day
- Thomas Jefferson University
- 2008 G Protein Signaling Workshop
- New York, NY
- 2005 Poster Penn State Summer Research Fair
- Pennsylvania State University,
Penn State College of Medicine
A current vitae is available in PDF format Download PDF
As can be readily inferred from the research
interests outlined above, aside from cell biological research, I have a
fascination with computer science. This is an intellectual itch that
ranges from algorithm design and artificial intelligence to data
science—I have outlined some of the projects below that have
served as a vehicle for these interests. While addressing fundamental
issues of computation, many of these endeavours serve to also improve
understanding and proficiency of programming efficient and fast code.
Computer Science Interests
- Algorithm design and optimization
- Implementation of Python and C++ in high(er) performance environments
- Artificial intelligence and the application to everyday problem solving and data analysis
- Machine learning for large data sets
- Making big data and computation available through web-based interfaces
Projects include
- Parallelization of pi computation in Python
- Google Code Jam and Project Euler in Python
- Conway's Game of Life with algorithm optimizations (cluster determinations)
- Weather data collection and analysis in MySQL and Python/BASH
- Laboratory resource and order management software in a custom Python web framework
- Big int factorization algorithms in Python and C++
- Mining scientific manuscripts for abbreviations with Python and Regex
Code and related projects can be found on Github
... Then felt I like some watcher of the skies When a new planet swims into his ken; Or like stout Cortez when with wond'ring eyes He stared at the Pacific ... - John Keats, ms of sonnet (1816)
1. Write down the problem. 2. Think real hard. 3. Write down the solution. -"The Feynman Algorithm" as described by Murray Gell-Mann

 In addressing questions of this sort, research evolves as a sort of biphasic
process. The first is the establishment of signaling pathways in the cell in
both the normal state and in response to environmental stimuli, and how these
lead to physiological changes. The second is how the cell detects and handles
aberrations to these processes to ensure fidelity. However, both phases require
the pursuit of a similar root understanding. Whether interrogating a typical
housekeeping pathway or a regulatory network attenuating an aberrant signal, the
cell must translate low level chemical changes to high level outcomes. It is not
surprising that cell biology is growing to encompass numerous fields of study
and exemplifies the emergence of truly multidisciplinary research.
In addressing questions of this sort, research evolves as a sort of biphasic
process. The first is the establishment of signaling pathways in the cell in
both the normal state and in response to environmental stimuli, and how these
lead to physiological changes. The second is how the cell detects and handles
aberrations to these processes to ensure fidelity. However, both phases require
the pursuit of a similar root understanding. Whether interrogating a typical
housekeeping pathway or a regulatory network attenuating an aberrant signal, the
cell must translate low level chemical changes to high level outcomes. It is not
surprising that cell biology is growing to encompass numerous fields of study
and exemplifies the emergence of truly multidisciplinary research.
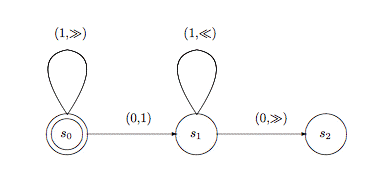 Addressing problems from a systems level requires an effective integrative
research approach, combining traditional biochemical and molecular biological
techniques with novel computational and microscopy methodologies. While adapting
new technologies for basic science research I will be further working towards
developing models of cellular processes and their regulation as computational
machines. The goal is to be able to establish the basic algorithms of these
networks and develop sets of cellular heuristics that comprise the decisions
that are undertaken. This algorithmic description of cells will further enhance
our understanding of cell biology while allowing for novel quantitative
approaches from a systems level.
Addressing problems from a systems level requires an effective integrative
research approach, combining traditional biochemical and molecular biological
techniques with novel computational and microscopy methodologies. While adapting
new technologies for basic science research I will be further working towards
developing models of cellular processes and their regulation as computational
machines. The goal is to be able to establish the basic algorithms of these
networks and develop sets of cellular heuristics that comprise the decisions
that are undertaken. This algorithmic description of cells will further enhance
our understanding of cell biology while allowing for novel quantitative
approaches from a systems level.